-
Posts
4586 -
Joined
-
Last visited
-
Days Won
12
Content Type
Profiles
Forums
Events
Everything posted by hypervalent_iodine
-
! Moderator Note It seems we are done here, so I’ve gone ahead and closed the thread.
-

Gregory Trovski's leap towards the "Theory of Everything"
hypervalent_iodine replied to Gregorytrovski's topic in Physics
! Moderator Note We ask that you provide enough information in the thread that members don't have to watch a video to know what you're on about. I am closing this. If you decide to reopen, please actually provide some text with a point of discussion. -
! Moderator Note Randall Canham, since it seems apparent you have no intention of being anything less than vague, and since this forum requires you to be precise and to support your ideas with proof and evidence, this thread is closed. You are not permitted to reintroduce it.
-
! Moderator Note Please return to the topic. OT posts will continue to be removed, and we will be adding official warnings and other sanctions if this continues.
-

Designing Primers for PCR
hypervalent_iodine replied to chesspuma99's topic in Biochemistry and Molecular Biology
I believe CharonY means that you can include the sequence for the digestion sites in the primers, which will then mean that those sequences are amplified along with the rest of the sequence and integrated into your PCR products. -
It's the natural log (ln) of 2. You can see it derived here: https://chem.libretexts.org/Bookshelves/Physical_and_Theoretical_Chemistry_Textbook_Maps/Supplemental_Modules_(Physical_and_Theoretical_Chemistry)/Nuclear_Chemistry/Nuclear_Kinetics/Half-Lives_and_Radioactive_Decay_Kinetics.
-
Right. I think this supports my suggestion of using your draw tool as a separate tool for student learning rather than the main one you use for all of your drawing. I think you might have misunderstood what I meant, or I wasn't very clear. I wasn't talking about generating those parameters on-the-fly, I meant that for your reaction solver or discover function it might be useful to be able to generate a structure from a substance identifier rather than having to draw it in every time. I question how robust this will really be. What you are describing is automated forward reaction planning, which has to rely in some part on automated retrosynthesis and / or the ability for your program deduce a way to build the right complexity to meet a defined end goal. Keep in mind that automated retrosynthesis is something that has only seen real-world success in the last year or two, and relies very much on AI / neural networks / deep learning / other buzzwords. What I am a little confused about is the ultimate goal of this website. The above quote seems to suggest you want it to be used for both researchers as well as students. If that is the case, I think you might be biting off more than you can chew with this particular tool. A research chemist seeking out synthetic protocols would have any number of concerns that I think you might struggle to address. For instance, what if I wanted to make something enantioselectively (this is a very common requirement)? Can it handle only linear synthesis, or could it develop a convergent route (the latter being much more preferable )? I can definitely see how handy it would be to have what you are talking about for a researcher if it were fully fledged and capable of handling a lot of complexity and a lot of reactions, but at the end of the day I would really question whether or not this is a goal you can reasonably achieve within the frame of a free-to-use web-based service with a small team behind it (I assume this is the case, apologies if not). Moreover, I wonder if by pitching this for students and researchers, you are spreading yourself a touch too thin and venturing into the realms of doing too much at once. Everything else in your website to me seems like it is geared largely towards education, so the reaction solver / design function seems a bit...out of place I guess. That's just my 2 cents. I haven't really seen anything in your website that would lend itself to this goal. Could you elaborate?
-
Yes, this is what I was talking about with the order of operations issues and was pretty much what I thought was going on. When you do it in the calculator, you should put your various terms in brackets as I did in my work-through. Operations in brackets are performed before other multiplication steps. Essentially what you were doing was what I have written in the first example (sorry about all the hand-written notes, I am just useless with LaTex): When you put the equation into your calculator, make it a habit to use brackets around everything (as I did in my calculation in my last post). I would be cautious about doing each operation separately and then combining the values, only because you get rounding errors this way and sometimes that can mean you fall outside the range of accepted answers in an exam.
-
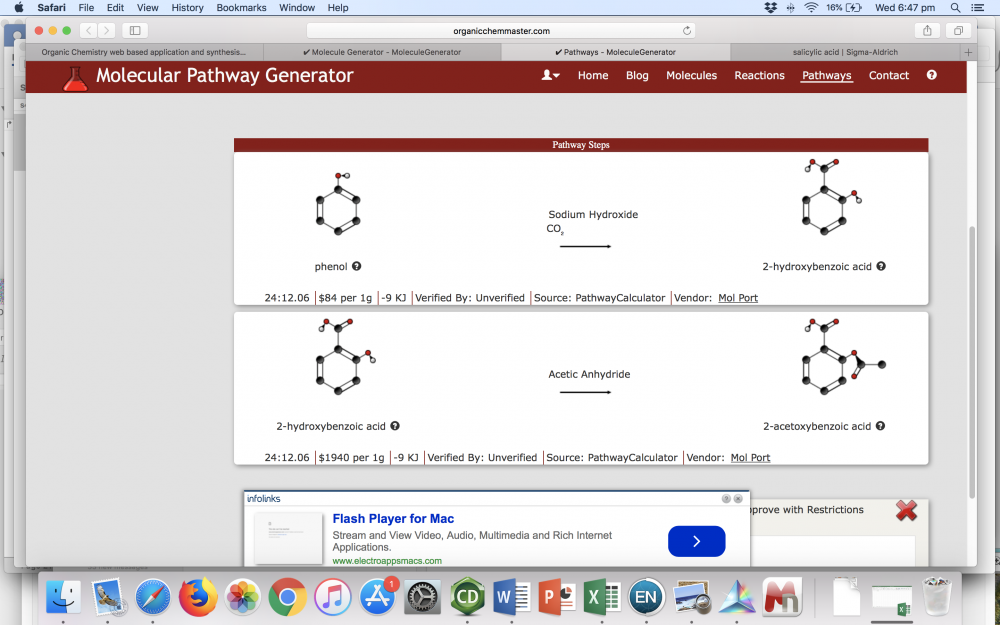
So, with Sigma you can get wildly different prices depending on what quality / grade you are okay with. For synthesis, you generally do not need analytical grade chemicals. I bring this up because when I went back to have a look at your reaction solver, I had a look at the suggested pathways for the example you linked to. This is what I get: The 2-hydroxybenzoic acid in the second example is aka salicylic acid. When I saw the price listed I immediately went and checked Sigma because there was no way something as common as salicylic acid costs $1940/g. When I check Sigma, I can buy 3 kg of the stuff for $US200. The only thing I could find close to the price you list was an analytical reference sample (50 mg for $US123), but no one is using an analytical standard to perform synthesis unless they have an absolutely wild disregard for budgets. One of the reasons I asked about where you were basing costs is because if you live somewhere like I do (Australia), you have to factor in a lot of other costs for delivery and tax. This is not really a problem if the intended users are students and not researchers of course. Firstly, I don't understand the bold statement. Unfortunately, the tool you have now is not very easy or intuitive to use (IMO), which I think should be a key focus over simply making something that is different. I think that the tool that you have with the on the fly IUPAC name generation would be good as a separate tool. As in, don't use it as the primary draw tool for your reaction solver or whatever, just use it as a specific tool for students to learn about IUPAC nomenclature. I think having something that creates a name as you build a molecule would be very good for that, but I can't think of a single time when I have been using SciFinder or similar to look up a reaction and have needed a IUPAC name to be generated as I construct a molecule. Another comment I would make about it is that you should integrate a way for a user to freely move around atoms. I was using the tutorial just before to make aspirin, and when I added an ethane to an oxygen atom, it added it in such a way that it overlapped with the phenyl ring. Upon trying to add the carbonyl oxygen, I instead got a carbon that appeared triple bonded to my original oxygen, and there was no way for me to change it without deleting it. I would also recommend you have a function to convert other identifiers into structures, like CAS numbers, SMILES, or IUPAC names. Finally, more atoms! Boron chemistry is pretty common for example, as are molecules with phosphorus, but you have neither as an option. Yes, you are correct. I half misremembered what it was and didn't look it up when I posted. Still an interesting project! From what I can tell, you are essentially automating a retrosynthetic analysis on a compound back to some pre-defined starting material, and then displaying that as the equivalent forward reaction steps. These forward reactions are shown as A --> B with the main reactants that one might use. Before I comment further, could you tell me if this is this correct? What level of complexity do you anticipate you will be able to handle? As in, how many retrosynthetic steps could it do, and how complex can the molecule be? I happen to work as an organic chemist in a group that does a lot of drug discovery and natural product total synthesis. In total synthesis, the things that we really aim for are (as I mentioned) shorter steps, higher yields, and fewer chromatography steps. The big thing is shorter routes and higher yields. Cost of the reagent is obviously critical also, particularly in industry, but having few steps, high yields, and easy purification are often the things you look at first.
-
Ahmed Torah has been banned permanently for posting a lot of preachy nonsense.
-
Never mind, I misread the post! I got the right answer by plugging your numbers into the equation. Are you using brackets around the exponential term? And are you then multiplying your values together? My suspicion is that you aren’t accounting for order of operations with the terms in your fractions. I’ve attached a quick scribble for how I got to the correct answer: I changed the units for delta G and R, because I don’t like kcal, but the numbers should still work regardless.
-
I don’t understand your first question. Or your second one, really. The Boltzmann constant is not k, it is kB (the B should be subscripted). In your calculation, you put the Boltzmann constant where k goes, which is incorrect. The value for k is the reaction rate constant, and this is what you are trying to solve. IOW, you treat k (the term on the LHS) as an unknown and solve the equation. This is what you have written in the picture you uploaded in the OP. From what I can tell, the only thing you were doing wrong there there was not using e correctly.
-
Are you confusing the rate constant with the Boltzmann constant? The LHS of the equation is k, which is the reaction rate constant (i.e. the value you're trying to calculate). The value you enter there is the Boltzmann constant, which belongs on the RHS of the equation (it is symbolised by kB). I would also suggest using brackets just to be safe you aren't getting caught by order of operations issues. I get s-1, so your math is out again. The Boltzmann constant is in J/K, temperature is in K, and Plank constant is J.s, so it looks like this: ((J/K)*K)/(J*s). The J and K units cancel, leaving 1/s.
-
Your calculator should have an ln / ex function. That's what you're looking for.
-

Notch1 receptor drug validation assay
hypervalent_iodine replied to haquee3's topic in Anatomy, Physiology and Neuroscience
Unless I have misunderstood, the OP is looking to perform a high throughout screening assay. A quick Google search suggests this has been done before, so you should be able to find a suitable protocol for identifying compound hits easily enough. See here, for example. -
Okay, I have looked at it briefly on my lap top. It is a good idea, but I think you need to invest a lot more time into it before it becomes useful. Before I get too much into it, I have to say that I don't like the draw tool you use. It's very limited and not super intuitive. I tried to step through the tutorial to draw ethanol, but could not get it to add any heteroatoms? Could you not integrate something like JSME or Chem Doodle? These are more complete in terms of a draw tool, and they use the more common bond-line / zig zag style of drawing. I think JSME is free to use as well. You may be interested in a recent project launched by IBM. It essentially performs the task of your reaction solver but uses a data driven AI approach, which I think is very elegant. Is this what your tool does as well, or do you use a different method? Out of curiosity, how do you calculate cost and reaction time? Does it take in to account purification and work up steps, or just the reagent cost (and if so, where do you source this and what country are you basing it from?). Reaction time is quite variable doesn't always translate across different molecules very well. I think most chemists would be more interested in limited number of steps, as this invariably leads to a reduction in cost and time. I don't think that trying to sort by some arbitrary cost or 'time' value would be that helpful. Another question I have with the solver feature is how it accounts for possible by-products and incompatible functional groups. Does it incorporate protecting group chemistry? Does it allow for the user to refine the types of reactions used? As for typos. Firstly, I would recommend sub-scripting in your chemical formulae where appropriate. You also use a lower-case k for K2Cr2O7. The wording you use is also very confusing in some places. I don't have time to go through it all, but I recommend hiring someone to proof read it. For example: This is the action for Cl2 addition to a double bond. I honestly do not understand what it is saying. Is it saying you only add one chloride to the double bond? That seems to be the interpretation, but it is incorrect if this is the case. I also don't like the use of the word replace. You use it when describing oxidation of alcohols to aldehydes, but this isn't really a good way of describing it IMO. Another thing I noticed is that your LiAlH4 reduction rules do not seem to account for esters. Will you have an encyclopaedia of name reactions?
-
It doesn’t seem to be well optimised for mobile devices would be my first comment. My second one is that I don’t really understand what the website is trying to achieve exactly? As a synthesis pathway engine it is not particularly useful as-is. From what I could see you only seem to be able to optimise from a very limited selection of molecules using biochemical pathways? Perhaps this is just because I am viewing it on my phone. The discover function again is very limited, and contains some typos and other errors.
-
What about the rest of the calculation?
-
I did the calculation using your values and get the correct answer, so I have to assume you aren't entering it correctly into your calculator. Could you possibly post a photo of how you are entering it?
-

Can anyone identify this ?
hypervalent_iodine replied to Mletcb's topic in Microbiology and Immunology
! Moderator Note We aren’t doctors. -
Maybe common where you are. I don’t think deliberately making the confusion is that common here, or maybe it’s just me.
-
Have I missed something? You're the only person who has said Austria?
-
Perhaps I am just cynical of our major parties and their ability to make decisions for any reason other than political point scoring, but I'll believe it when I see it. He has stated as recently as the past week that he supports keeping the date as-is.