
fredreload
-
Joined
-
Last visited
Posts posted by fredreload
-
-
-
-
Alright, so you got an object going near the speed of light and he perceives the universe with a length close to zero like a contour around the object, but what is outside of this contour? What lies beyond this universe that is being rigored and everted? The future block universe? And once this future block universe gets everted by your perceived reference frame, what happens when you attempt to enter this everted universe? Will you get spits back out?
-
-
-
Two object, yellow and red, is traveling close to the speed of light 3*10^8m/s, and the black circle is the space time that they perceive around them.
1st question. Are the two space time perceive by yellow and red two distinct space times?
2nd question. If they pass through each other(as the arrow shown), does the space time maintain their original identity after they pass through each other?
-
-
19 hours ago, swansont said:
Modulation on a wire would be radio, if the signal is RF. It would not produce visible light.
Yes, your logic is correct. You do not need an alternating current to produce light from tungsten.
18 hours ago, studiot said:Since you evidently don't wish to talk to me I will bow out.
Hi Studiot, I am just curious on the data modulation from an electrical current onto the radio wave. Reading a 200 pages paper for this might be a bit too long.
-
Edited by fredreload
Can you apply an alternating current on tungsten to produce light wave for communication? I am trying to mimic how radio wave works, which uses alternating current on an antenna.
P.S Seems like someone got it to work already @@, respect. https://en.wikipedia.org/wiki/Li-Fi
-
-
-
-
So I was looking at this video about Tesla turbine and I came up with a new idea. What if we use a magnetic field, like a motor, and increase its speed inside a vacuum with 0 gravity acting on its friction so it just keeps spinning faster and faster? Well I think one of two things would happen, either the material would not hold(breaks apart), or a whole hole be created in space time, leading to a previous, next block of the universe. Let me know what you think.
-
Edited by fredreload
55 minutes ago, swansont said:That’s how that imager works
Your link describes multiple imaging methods. You should read it more carefully.
“The moving scanner line in a traditional photocopier (or a scanner or facsimile machine) is also a familiar, everyday example of a push broom scanner.”
Maybe you could read the links you post
About the video I posted regarding thermal imaging. The idea is to have a "sensitive sensor" capable of trapping and reading infrared wavelength(That is where the CCD camera comes in). An analogy would be the eye and the retina and how we see visible light. To answer your question, a light wave does not exhibit a 3 dimensional structure, but the timing does.
https://midopt.com/solutions/monochrome-imaging/infrared-imaging/
-
5 minutes ago, swansont said:
Did you read you own wikipedia link?
You need multiple pieces of information per pixel to get the image. The link explains ways to do this.
Meaning your understanding is incomplete
See there is no slit in this one = =. I think the lined scan idea is metaphorical to the data array of the greyscale camera. How about the second question?
-
Edited by fredreload
5 minutes ago, swansont said:It does what?
Not unless they are transparent at the wavelength in question.
You ignore the third dimension, or incorporate the info in the image. You do realize the 3rd “dimension” is wavelength, right?
“There are four basic techniques for acquiring the three-dimensional (x,y,λ) dataset of a hyperspectral cube.”
In this sense a regular photo is a 3D image.
Here this video says that the hyperspectral imager requires a line scan. I mean does that slit contains the information of the entire leaf or do you need to scan it? And a thermal image camera does not require a line scan I think? This does not add up.
-
Edited by fredreload
So first thing first, I took a look at the hyperspectral imaging page from Wikipedia(https://en.wikipedia.org/wiki/Hyperspectral_imaging) and as most of you know it is s system that utilizes Raman laser(laser reflection) and spectrometer. Now what is interesting is this time I found an application call thermal imaging in the same page in the Wikipedia page produced by a Specim LWIR hyperspectral imager.

Now thermal imaging is quite common that utilizes the infrared light. Now the question is I am unsure on is how the hyperspectral image is produced because technically the infrared light goes through objects, so if I want to see through a person or a car I would have to utilize a time scale, possibly with the Fourier transform equation. So this is my first question on how they obtained a 2D image from a 3D imaging system. Below is a link to the Specim hyperspectral imager.
https://www.specim.fi/downloads/Spectral_Cameras_LWIR_ver2-16.pdf
Now the second question is I want to combine this hyperspectral imaging system with a zoom in effect. Now as you know if I want to magnify an object I would use a lens, but for infrared I might need a different material for the lens since the wavelength is different. But in the end a CCD camera records the spectra of the sample, so if the equipment is sensitive enough, I might not have to magnify the infrared wavelength of light using special infrared optic material. This is question number 2. Below is a link to the ccd camera.
https://www.tandfonline.com/doi/abs/10.1080/10106040308542274
On further investigation I found this (https://en.wikipedia.org/wiki/Push_broom_scanner). If someone could explain to me how it works.
-
-
Edited by fredreload
On 5/29/2021 at 5:07 AM, Sensei said:No. To create a current, a photon would have to be absorbed by an electron which then would be ejected from the medium's atoms and accelerated. The ejection of an electron from an atom in a medium requires the addition of energy.
Such thing happens in e.g. photoelectric effect:
https://en.wikipedia.org/wiki/Photoelectric_effect
A photon passing though a transparent medium has to exit it from the opposite side (otherwise the medium is opaque).
An opaque medium can: reflect photons, refract photons (in random directions) or absorb photons.
(a transparent medium for photon with one energy/frequency/wavelength is often opaque for photon with different properties!)
An alternative (and actually more common) thing that can happen after a photon is absorbed as it passes through a medium is heating it.
Radiowave antennas are absorbing some photons, which accelerate some (a very few) electrons in the metal, in one direction, then in other direction (depending on photon properties). Therefore the antennas are connected to amplifiers https://en.wikipedia.org/wiki/Antenna_amplifier which detect these minor currents and amplify them.
Can you induce photoelectric effect with ultrasound?
P.S. Alright, it is not photon, but how does an ultrasonic transducer convert ultrasound to electrical current?
-
So I found this microwave antenna unit on Alibaba. I want to turn it into an MRI machine to image the brain. How should I go about working on it?
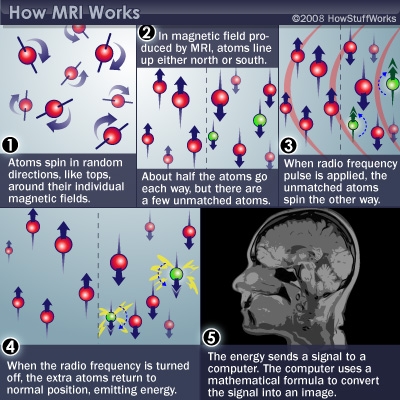
Following this chart based on my speculation:
1. Apply microwave on the brain to fix the atoms(water molecules) on a correct position since the microwave contains a magnetic field.
2. Apply a separate microwave pulse to turn the unmatched atoms and receive the reflected microwave.
3. Construct the image based on the microwave signal.
The microwave antenna Unit + Price:

How MRI works:

-
-
-
Edited by fredreload
5 hours ago, Sensei said:No. To create a current, a photon would have to be absorbed by an electron which then would be ejected from the medium's atoms and accelerated. The ejection of an electron from an atom in a medium requires the addition of energy.
Such thing happens in e.g. photoelectric effect:
https://en.wikipedia.org/wiki/Photoelectric_effect
A photon passing though a transparent medium has to exit it from the opposite side (otherwise the medium is opaque).
An opaque medium can: reflect photons, refract photons (in random directions) or absorb photons.
(a transparent medium for photon with one energy/frequency/wavelength is often opaque for photon with different properties!)
An alternative (and actually more common) thing that can happen after a photon is absorbed as it passes through a medium is heating it.
Radiowave antennas are absorbing some photons, which accelerate some (a very few) electrons in the metal, in one direction, then in other direction (depending on photon properties). Therefore the antennas are connected to amplifiers https://en.wikipedia.org/wiki/Antenna_amplifier which detect these minor currents and amplify them.
Yes, the antenna uses an AC current to create radio wave. Right, you are correct, the electrons does not appear out of thin air unless you can use a high enough voltage to zap out the plasma. In this case I might need to use electron doping to have the electrons surf along the infrared laser. Photoelectric effect requires visible light wavelength to have enough energy right? Or would infrared works?

-
1 hour ago, swansont said:
Just trying to fix your (quite egregious) errors on the topic in question.
You aren’t going to solve anything without understanding the underlying science.
Note that this is 1) in a medium and 2) does not alter the path. (Just in case fredreload is tempted to take Faraday rotation as confirmation that their assertion has any traction)
I see, I think Sensei is trying to hint that a rotating magnetic field would create current. Although creating a long range magnetic field is unheard of, but there is a device called transcranial magnetic stimulation based on electromagnet. Personally, I would probably use a high power infrared laser if I want to image the brain.
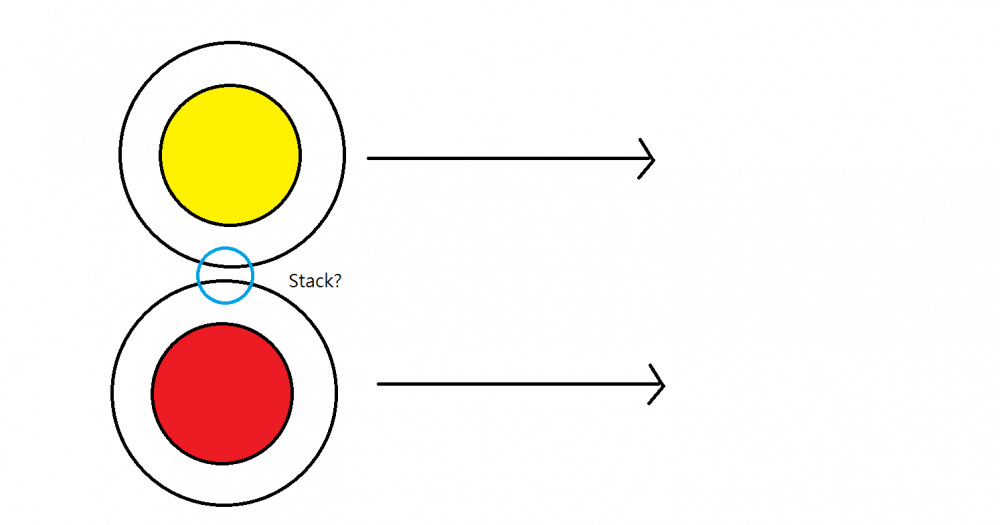
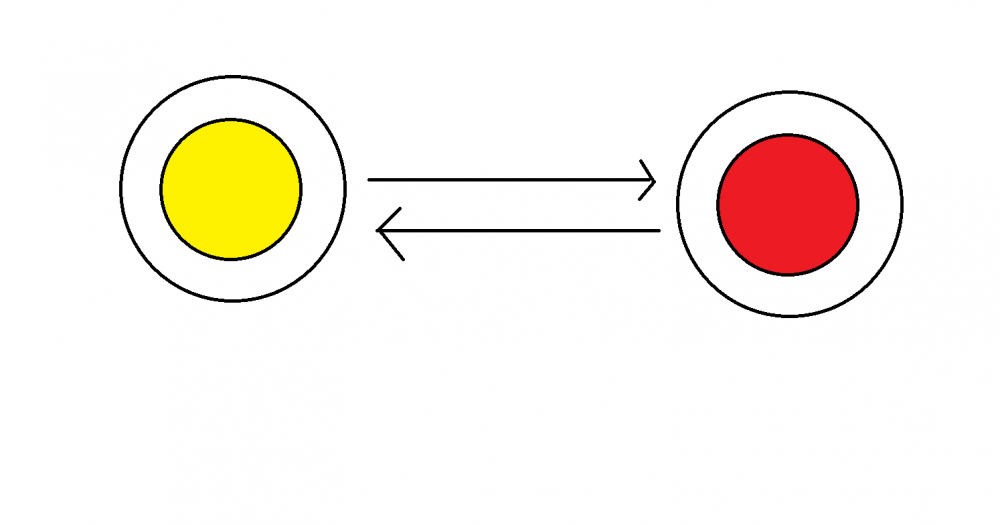
Two objects going [near] the speed at light(Relativity)
in Speculations
Let us see. You said you cannot evert the object by itself, but if you use the rigor method you will be left with an umbilical cord and therefore unable to connect to the future space time. The only way is if you rigor the future block space time as a separate non-connected instance to the present block of space time. If you attempt to sever the umbilical cord(An analogy of the lingering space time after sphere eversion.), then you could just cut through the present space time without sphere eversion(Which is still an unknown method.).