-
Posts
18 -
Joined
-
Last visited
Content Type
Profiles
Forums
Events
Everything posted by Micro.Pete
-

How do you prepare a pure bacterial spore suspension?
Micro.Pete replied to stbell's topic in Microbiology and Immunology
Yes and decided to answer anyway. If you don't want people to answer just close the thread.- 12 replies
-
-1
-

How do you prepare a pure bacterial spore suspension?
Micro.Pete replied to stbell's topic in Microbiology and Immunology
Yes and decided to answer anyway. If you don't want people to answer just close the thread. -

How do you prepare a pure bacterial spore suspension?
Micro.Pete replied to stbell's topic in Microbiology and Immunology
Have you tried using old Bacillus colonies (over 3 days old)? From my experience, Bacillus spp. sporulare on the agar after 3 days fue to nutrient deprivation. Just wait 3-4 days and check the old colonies for endospores, I'm sure you'll see over 90% endospores. Another thing, you don't always have to special stain your cells to see endospores. Just use Gram staining. After you Gram stain the cells you will see the endospores as a white area inside the bacterial cell (which stained Gram-positive). -

Help finding Streptococcus phages
Micro.Pete replied to N2Microbes's topic in Microbiology and Immunology
Have you tried writing to Eliava institute in Tbilisi, Georgia? The microbiology institutr is the world's leading research center on phage therapy since 1930s They have a lot of field and medical experience about phages, so try asking them. Here is the site of the institute: www.eliava-institue.com -

What could this organism be ?
Micro.Pete replied to Thaslim.mbio's topic in Microbiology and Immunology
My first guess would be a Serratia spp. There are a lot of bacteria in the genus Serratia and they can differ one from another (e.g. different colony pigment). You should note that not all Serratia species produce the well known red-pink pigment. Moreover, some species that usually produce it (e.g. S. marcescens), in some condition stop producing it. But this is of course only a guess. Further biochemical/molecular ID of the bacteria is needed. -

Enterohemorrhagic Escherichia coli O157:H7 Help!!
Micro.Pete replied to docjo.chua's topic in Microbiology and Immunology
Hi there, What is your question exactly? - EHEC differs from other E. coli strains being unable to ferment sorbitol, thus it uses peptone in the agar to grow and make colonies. The use of peptone raises the pH in the medium,. So EHEC will grow on MacConkey sorbitol agar producing pale to yellow colonies. -
Hi there, My guess is that your fixation wasn't perfect. Smear your bacteria on a microscope slide and wait until it is dry (you can put your slide in an incubator to speed up the drying process). After that you have to fix the bacteria to the slide. The easiest way to do it is to place the bottom side of the slide under a flame, such as a lighter or a bunsen burner. Just Make sure you don't burn your bacteria (the whole process should be quick). Hope that I helped.
-

Treating strep or not?
Micro.Pete replied to Marshalscienceguy's topic in Microbiology and Immunology
As many infectous diseases your immune system can and will ultimately clear Streptococcal infection, though it can take some time. However you should remember that in some patients and especially in children Streptococcal infection can cause some serious complication such as: scarlet fever, middle ear infection (Otitis media), post Streptococcal Glomerulonephritis, pediatric autoimmune neuro disorders associated with Streptococcal infections (PANDAS), rheumatic fever, endocarditis, and more. In most of the cases Streptococcus pyogenes (group A Streptococcus), and gr. C+G Streptococci are sensitive to penicillin drugs (e.g augmentin). They often seen as just Strep throat bacteria, while at the same time can cause other complications. -
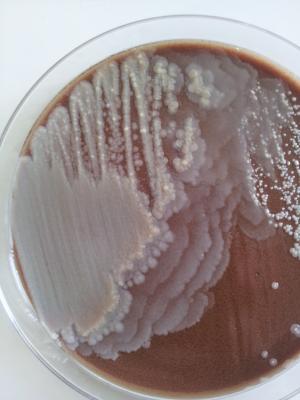
Many pathogenic bacteria have swarming motility (e.g: Bacillus spp., Serratia spp., Salmonella spp., Proteus spp., and many more). In pathogenic context the ability to swarm over surfaces allows bacteria to move, colonize and eventually invade tissues, cells and organs. Because of that, this ability gives those bacteria an advantage over non-swarming species. Thus your hypothesis sounds logical and plausible to me. And here is probably one of the best known swarming bacteria that exist: Proteus mirabilis. Seen here swarming over Chocolate agar. This culture was obtained from an ear swab.
-

Trouble shooting for Modified Hodge test?
Micro.Pete replied to vivegaviji's topic in Microbiology and Immunology
Hi, Please specify what is exactly your problem with the MHT. It can be due to the ATCC E. Coli strain or with the Klebsiella controls or even because of bad equipment (such as agar plates, saline, etc..) Make sure that you do the next steps: 1. For obtaining a good ATCC E. Coli growth you have to reach 0.5-0.63 McFarland dilution. For that you have to use extra pure saline (not the one you may use for the VITEK machine). 2. Make sure that your Mueller-Hinton agar plates are not expired. 3. Make sure you use fresh Klebsiella positive and negative controls (They are typically good for about 2-3 weeks after you prepare them on Mcconkey agar plates). 4.what kind of bacteria do you often check with your MHT? Note that Enterobacter spp. tend to give false positive results which have to be verified using PCR. -

Which test can I use to identify Bacillus cereus?
Micro.Pete replied to countessalyssa's topic in Microbiology and Immunology
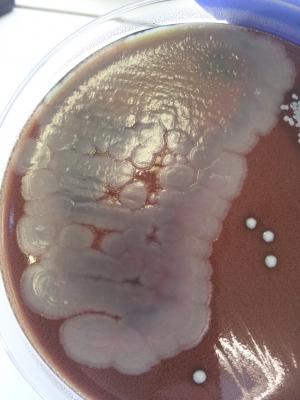
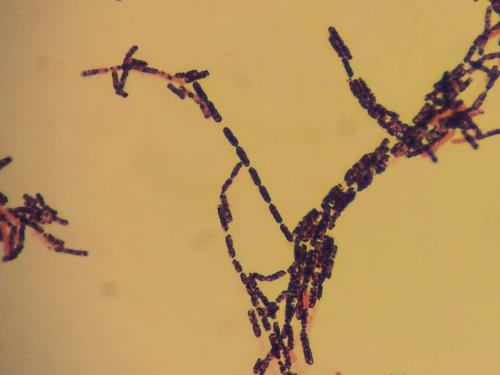
A good way to identify Bacillus spp. is by using API 20E, and 50CHB kits. Another way is to insert a special Bacillus card into the VITEK rapid microbial identification machine. Surelly the best way is to use mass spectrometer, but if can't use non of these, you will have to identify the bacterium by biochemical methods. Here is something that can help you: Bacillus cereus is: * Gram positive (but not always. Old bacteria in a culture can be stained Gram-variable) * Motile * Beta haemolytic * Facultatively anaerobic * Rod diameter under microscope is 1.2μm * Bacteria tend to form chains * Hydrolyzes casein, starch and gelatin * Reduces Nitrate * Doesn't produce indole * Doesn't produce gas from Carbohydrates * Produces acid well from Trehalose * Grows well on various agars such as: TSBA 5%, CNA, MRSA/MSSA CHROMagar, Chocolate agar. You should also note that despite these biochemical properties sometimes it's quite difficult to distinguish it from other Bacillus speciesת especially: B. amyloliquefaciens and B. thuringiensis. * First picture shows B. cereus colonies swarming over Chocolate agar. * Second picture shows B. cereus rods forming chains (X1000). This bacterium was isolated from an ear infection.- 1 reply
-
2
-

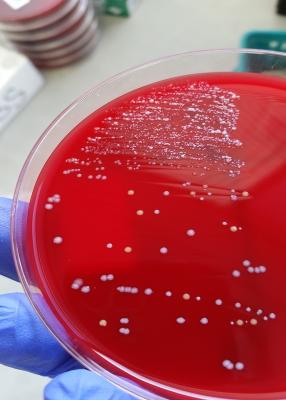
Where exactly to find M. Luteus?
Micro.Pete replied to assanater601's topic in Microbiology and Immunology
Today I have noticed several M. luteus colonies on TSBA 5%. You can easily spot the bright yellow bacteria among coagulase negative Staphilococci, which are white (see attachment). This culture was obtained from a vaginal swab. -

Medical student on microbiology
Micro.Pete replied to RodrigoCF's topic in Microbiology and Immunology
You can use different agar plates for isolating well known, and of course unknown bacteria. Than, after you obtain a clean-isolated colonies you can start working with them (e.g: Gram-staining, performing biochemical tests, picking colonies for MS analysis, etc...). -

Where exactly to find M. Luteus?
Micro.Pete replied to assanater601's topic in Microbiology and Immunology
As mentioned above, M. luteus is part of the normal human flora. In our medical microbiology lab we isolate it very often, especially during MRSA screening tests because it also dwells in the nasopharynx and as mentioned above colonizes all the respiratory tract. M. luteus grows well and fast (you can see its colonies after a period of 24 hours incubation) on TSBA and CNA agars, as well as on Chocolate agar. As its name indicates, the bacterium has a yellow pigmentation which you can easily notice while observing the colonies. I must add that it is being often confused with Staphylococcus aureus colonies, mainly because of their pigmentation. But while S. aureus has a golden or slightly yellow pigmentation, M. luteus has a bright yellow one. * As you may already know, S. aureus is also part of the normal human flora (~80% of the population carry it in the nasopharynx. so you can easilly isolate them both), so in order to be 100% sure that you deal with M. luteus and not with S. aureus I advise to do the following: 1.) Compare their pigmentation as noticed above. 2.) Gram-stain the suspected colonies: S. aureus (and also other Staphylococci) is a Gram positive coccus which clusters to form a grape-cluster shape, while M. luteus is a Gram positive coccus which tends to form quadruplets. 3.) S. aureus is coagulase positive and DNAse positive, while M. luteus isn't. Hope I helped! -
It is an unusual question, but I think an interesting one. I'll try to answer it scientifically: Here are some facts we all know: * B. cereus is a endospore forming bacteria. * S. aureus has a natural yellowish-golden pigment (Staphyloxanthin). * Some strains of S. mersescens have a redish-orange pigment (Prodigiosin). * S. cerevisiae yeasts have a white pigment. Bacterial endospores are highly resistant to the environmental conditions, such as: radiation, temperature, pH, humidity, etc. We all know that carotenoid pigments on animal and human skin protect them from UV. the same we can say about bacteria. Gram positive bacteria have thicker peptidoglycan cell wall than Gram negative bacteria. So if I had to answer the question, I'll probably place the organisms in the following order of sensitivity to UV: (Less resistant) Saccharomyces cerevisiae>Seratia marscescens>Staphylococcus aureus>Bacillus cerreus (Most resistant).
-

antibacterial activity via disk diffusion assay
Micro.Pete replied to ken77's topic in Microbiology and Immunology
Hi, If you find a haze around your DD, and it appears to be bacteria (try to scrub it with a inoculation needle), it means the antibiotic isn't working against this microorganism.That applies only when you work with a clean isolate (when you have only 1 bacterial species). Because if you work with a mix culture, you can't be certain in 100% that the haze came from your tested bacteria or from other species in the mix culture. -
Hi Tentacle, I'll try to answer your questions: As you probably may know the Human papillomavirus can infect keratinocytes in the skin and also mucous membranes in the body. In fact the majority of HPV infections occur in the skin and are sub-clinical (without any symptoms). One way to transmit HPV, rather than during sex, includes infected skin of the hands. Thus by infected hands one can transmit the virus to other parts of the body, such as the genitals. Transmission of the virus using contaminated equipment is possible, but is less common and quite rare, so I really think the main reason for the infection was caused by hand-genitals contact. Note also the not all the HPV strains are carcinogenic. From about ~170 HPV types that were identified only 15 are carcinogenic (among them types: 16, 18, 31, 45, are placed under the "high carcinogenic risk" group). The best thing to do is to continue the regular Pap test screening. She can also ask her physician to take a genital swab and send it to the virology/microbiology lab for HPV detection and typing.
-
Hi all, I have a question regarding acterial isolate. Several days ago we recieved a sawb from a patient with an eye infection. After a period of 24h incubation on Chocolate and TSBA agar I have noticed stange looking colonies. The colonies were small, white and creamy and resembeled coagulase negative Staphylococci (e.g; S. epidermidis), which were also present on the agars. So I Gram-stained the bacteria and in parallel prepared a sample for Maldi-tof MS. The Gram staining revealed Corynebacterium spp., and the MS analysis was Corynebacterium pseudodiphtheriticum. My question is whether this bacterium is known to cause eye infections, or is it a part of the normal flora (as other non-Corynebacterium diphteria species are). Thanks,