Spezialemic
Members-
Posts
13 -
Joined
-
Last visited
Content Type
Profiles
Forums
Events
Everything posted by Spezialemic
-
Hey guys, So I'm trying to do a diazotization of 3-aminopyridine (as the title has no doubt tipped you off), and I'm having some trouble. I'm dissolving the pyridine in excess of water, adding 1.2 env HCl and cooling the water in the flask with an acetone/dry ice bath down to -5ºC. I add NaNO2 in water dropwise to the flask but I don't see any redness or evolving gas, as I would expect to (so my PI says). After addition and enough time for homogenization, I reflux at 70ºC for several hours. The product does not form. Any ideas?
-
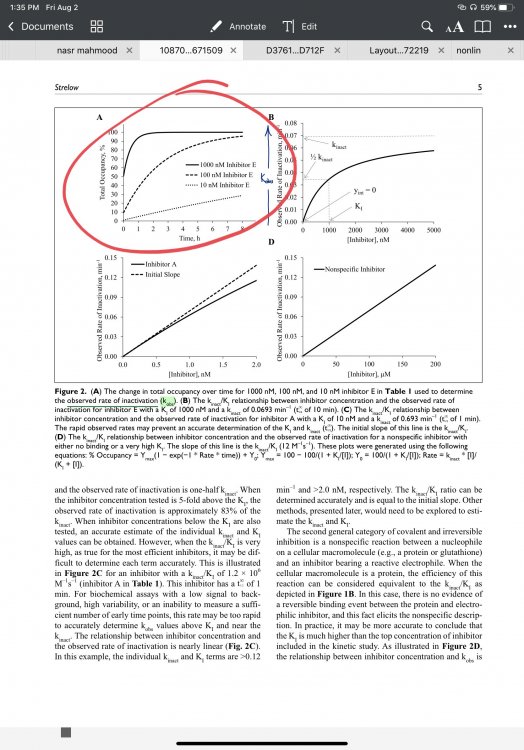
I’m doing a study to determine the rate of covalent modification, also known as kinact/Ki. From the literature, I see that it is determined by plotting percent occupancy over time (DOI: 10.1177/1087057116671509). I ran a 3 pt (8 compounds) fluorescent assay over 180 minute and normalized the data to the blank at time t and the final positive control. This activity vs time was converted to percent occupancy over time with a typical (1-value)*100. The graphs this generates are not consistent with what is expected from the literature, and I’m curious as to whether I have done something incorrectly, or if these compounds are not, in fact, covalent; I’m not sure how likely that is. I have included the spreadsheet. Keep in mind if you backtrack the data that E,G, and H9 and all column 10 values from the plate were excluded due to the availability of the substrate. Because of this, looking at values besides those in compounds 5, 7, and 9 are most representative of the proper formatting. Any help would be greatly appreciated! Thank you Kinetic Study Post.xlsx
-

Difficulty with maleic anhydride compound
Spezialemic replied to Spezialemic's topic in Organic Chemistry
No work up. Just evaporation. -

Difficulty with maleic anhydride compound
Spezialemic replied to Spezialemic's topic in Organic Chemistry
I can post the NMR on here a bit later. The problem is it’s still a very crude product. -
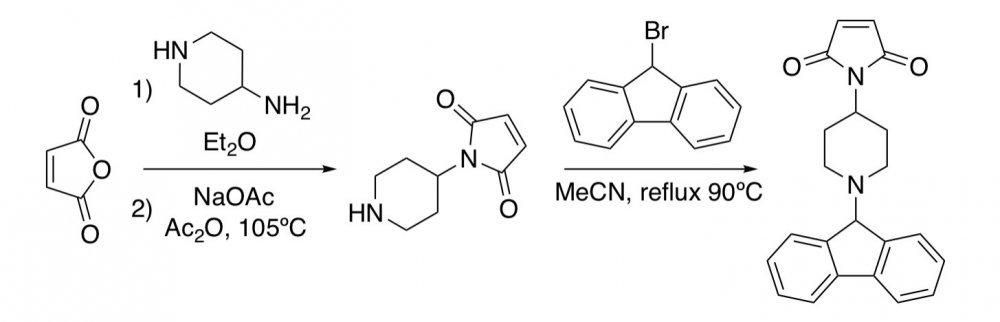
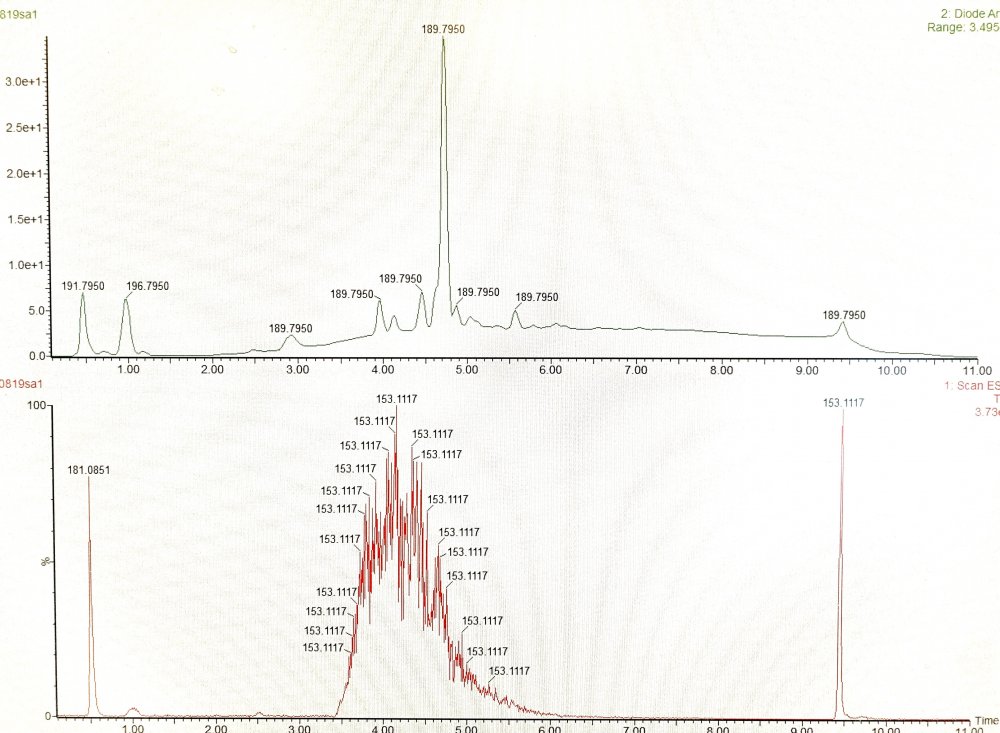
I’m having a little trouble with a reaction and was hoping someone could help me. I’m trying to make a maleic anhydride compound, but the compound is not there in the final step. My procedure is as follows: To a clean and dry RBF fitted with a reflux condenser and a 1” stir bar, 20mL of dry ether and 0.004mol maleic anhydride are added. The mixture reflexes at room temperature under argon until the maleic anhydride is dissolved, at which point 0.004mol 4-aminopiperidine in 8mL dry ether is added. A white solid immediately precipitated, and the mixture stirs for an additional hour before being evaporated under reduced pressure. The resulting white powder was determined by LCMS to be the correct mass for a mono-substituted piperidine (thought the position is merely assumed to be at the primary amine). Next, the product is dissolved in 20mL acetic anhydride and 0.004mol sodium acetate is added along with a 1” stir bar. The reaction mixture stirs in a 105°C oil bath for 30mins, over which time is adopts a dark brown color. The mixture then cools in a water bath for 30 minutes before be evaporated under reduced pressure. The resulting brown powder is dissolved in 20mL acetonitrile and 0.004mol 9-bromofluorene is added along with 0.0048mol DBU and 0.02mol Sodium Bicarbonate. The reaction mixture reflexes at 90°C in an oil bath for 40h and is evaporated under reduced pressure. The LCMS for the final crude is included. As you can see, the product did not form and it just appears that there is a lot of DBU. Does anyone have any ideas for a better path to take for this reaction?
-
I'm trying to do the reaction shown here, but when I perform it I simply get two spots after w/u with DCM/H2O which are masses 213 and 251 (both +/- 1). sans an NMR, what might these two products be and how could I prevent their formation? Any ideas? Thanks!
-
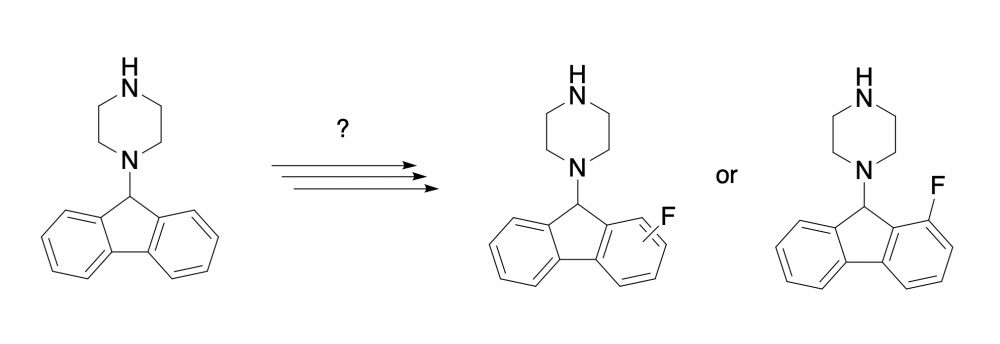
Hey guys, I need to fluorinate a fluorenyl piperazine fragment for an NMR experiment I'm running, and I'm not quite sure what the best way to go about it would be. I'm striving for the uni- or bilaterally fluorinated product preferably at the 1-position (or 1 and 8, bilaterally) for reasons related to protein binding sterics. However, fluorination at any of the 1-8 positions, either uni or bilaterally would still be acceptable. I'm asking here since scifinder hasn't been able to lead me in the right direction so far, so I'm hoping someone here might know where to start. Thanks!
-

Impure 9-bromofluorene from Sigma, but issues purifying
Spezialemic replied to Spezialemic's topic in Organic Chemistry
What do you think of the most recent NMR, though? It seems like the problem resolved itself. I wonder if my chloroform D was contaminated -

Impure 9-bromofluorene from Sigma, but issues purifying
Spezialemic replied to Spezialemic's topic in Organic Chemistry
EtOAc and Hexane -

Impure 9-bromofluorene from Sigma, but issues purifying
Spezialemic replied to Spezialemic's topic in Organic Chemistry
So I re-ran the NMR in acetone-D6 1% (v/v) TME and those peaks disappeared, replaced by the acetone, HOH, and HOD peaks, I presume. I think my CDCl3 might be contaminated. I also notice that the expected peaks are shifted more downfield in acetone. -

Impure 9-bromofluorene from Sigma, but issues purifying
Spezialemic replied to Spezialemic's topic in Organic Chemistry
I haven’t done a 13C yet because there hasn’t been a long enough block of time I could grab. Our LCMS is also down until the new column comes in cause someone just ran a really concentrated sample the other day. I wouldn’t be surprised if those are from other solvents, though. My vacuum pump needs to be opened and cleaned, so I’ve been using the hood vac. -
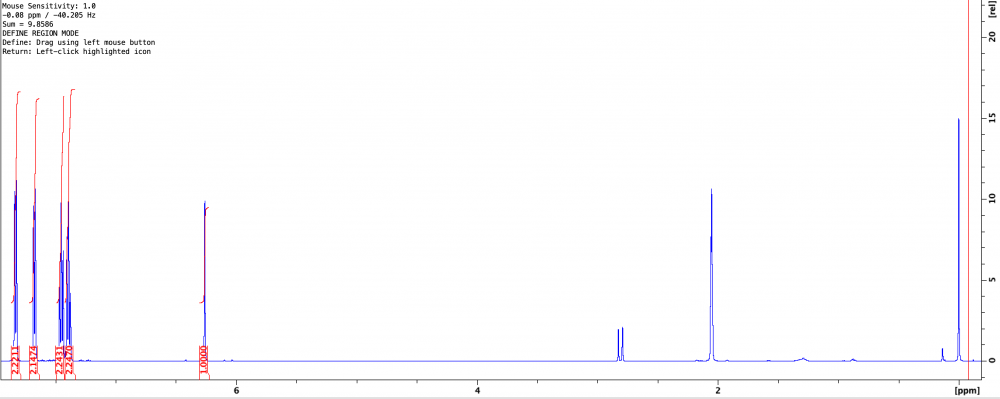
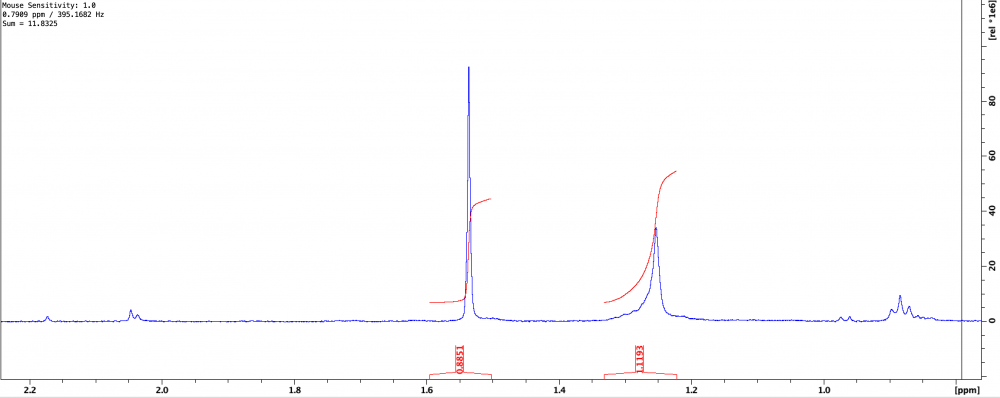
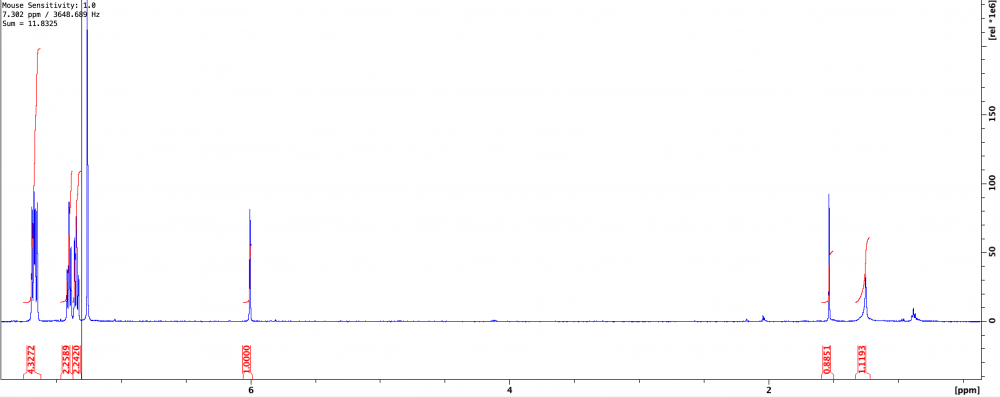
So I had a stock of Sigma 9-bromofluorene I planned to use to generate some new chemical antagonists. Problem is that the TLC showed 4 spots. Ordered fresh stock, still came impure. Issues with Sigma aside, I need the compound so I just ran it through the biotage...TWICE...and it’s still not completely pure. The second purification did yield only one spot, but the NMR was not clean and being a student I’m not familiar enough with spectroscopy to determine where these peaks upfield are coming from. I’ve included the NMR (CDCl3) and maximization of the troubling peaks at 1.25 and 1.5. Though I’d usually assume 1.5 to be water, this was after a flash purification in 1% EtOAc:Hex, and a nice long rotovap, so I don’t buy that it’s a water peak since it’s so intense. I’m thinking everything upfield of 1 is just hexane. The TLC still shows only one spot and most of the impurities (if not all) are gone, but I’m hoping someone can help.